
Peter Schubert, Prof. Martin Lercher and the team of the Institute of Computational Cell Biology, Heinrich-Heine-University Duesseldorf, Germany
Welcome to sbmlxdf’s documentation!
Convert between SBML and tabular structures
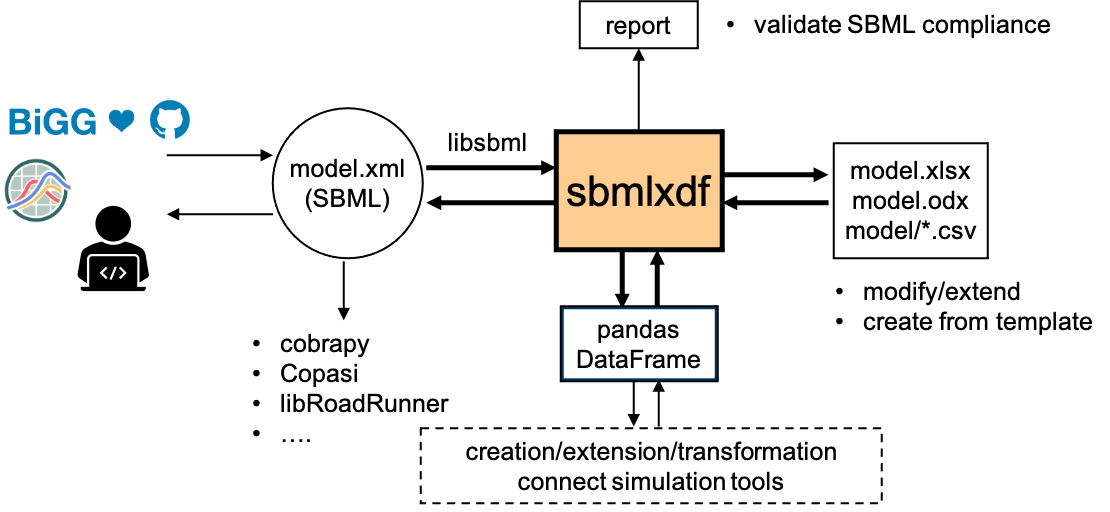
sbmlxdf is lightweight and transparent converter from SBML to pandas Dataframes (sbml2df) and from pandas Dataframes to SBML (df2sbml).
sbmlxdf supports, with few exceptions, all functionality of SBML L3V2 core package [SBML_L3V2] and packages Flux Balance Constraints (fbc) [SBML_fbc], Groups (groups) [SBML_groups] and Distributions (distrib) [SBML_distrib].
At the backend, libSBML API is used for accessing SBML data elements [libsbml].
Benefits
kinetic modelers with and without programming skills
overcome hesitation of creating own models in SBML
have a tool for flexible kinetic modelling using spreadsheets
inspect SBML models
create/extend SBML models
use latest SBML features
generate ‘correct’ SBML models
Python programmers
get access to SBML model data via pandas DataFrames, e.g. for use in their optimizers
can evaluate different model design strategies
Features
support of SBML L3V2 core [SBML_L3V2], including:
model history, miriamAnnotations, xmlAnnotations
units of measurement
local/global parameters
function definitions
Flux Balance Constraints package [SBML_fbc]
Groups package [SBML_groups]
Distributions package [SBML_distrib]
Note
sbmxdf does not intent to support SBML packages related to graphical representations of network models. I.e. packages Layout and Rendering are not supported. Other released SBML packages as of July 2021, see package status i.e. Hierarchical Model Composition, Multistate and Multicomponent Species and Qualitative Models are not supported at the moment, but might be included in future versions.